.ipynb
Create Custom Instance Segmentation Table¶
Create a 3LC Table from custom RLE annotations using the Sartorius cell instance segmentation dataset with hundreds of cell instances per image.
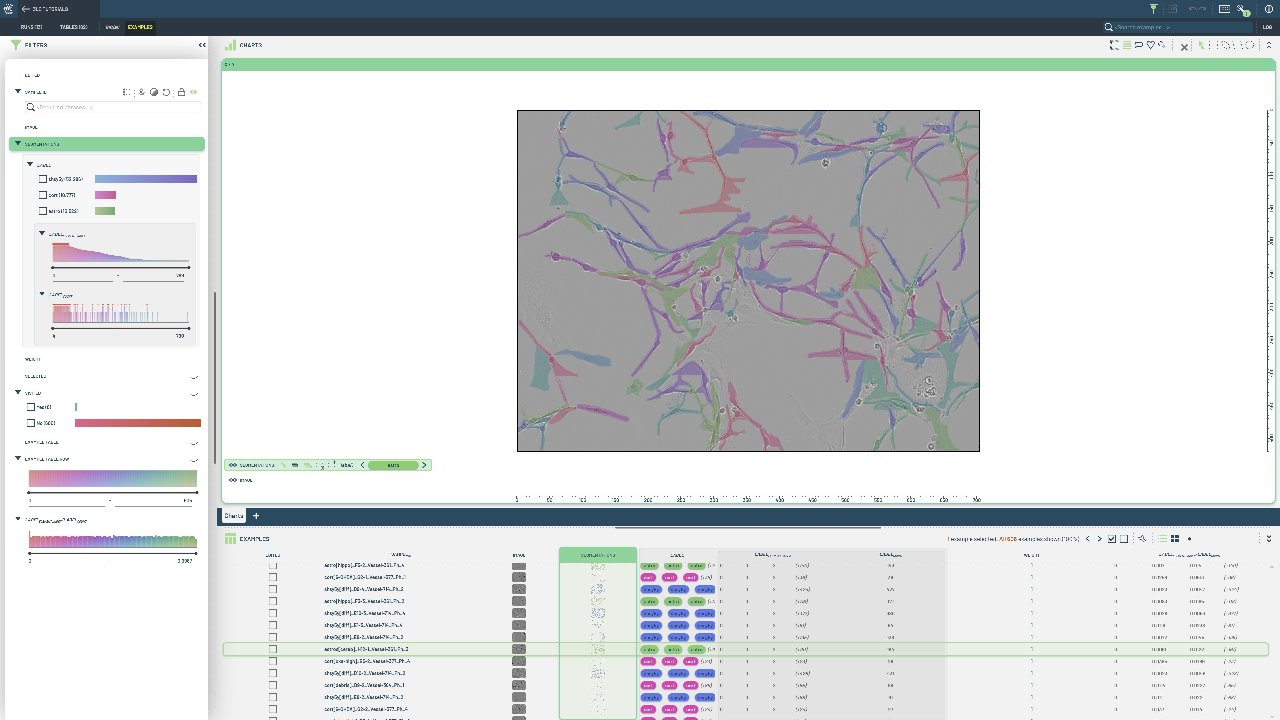
Custom instance segmentation is needed when working with specialized domains like medical imaging, where standard datasets don’t capture the specific characteristics of your data. RLE format is memory-efficient for dense segmentation masks.
This notebook processes the Sartorius Cell Instance Segmentation dataset from Kaggle, converting RLE-encoded masks to 3LC format. We handle multiple instances per image and demonstrate custom schema definition for specialized annotation formats.
Project Setup¶
[ ]:
PROJECT_NAME = "3LC Tutorials - Cell Segmentation"
DATASET_NAME = "Sartorius Cell Segmentation"
TABLE_NAME = "initial"
DOWNLOAD_PATH = "../../../transient_data"
Install dependencies¶
[ ]:
%pip install -q 3lc
%pip install -q matplotlib
%pip install -q kaggle
%pip install -q git+https://github.com/3lc-ai/3lc-examples.git
Imports¶
Prepare Dataset¶
The data needs to be downloaded from Kaggle before it can be used.
Either ensure you are logged in to Kaggle and the file ~/.kaggle/kaggle.json exists, or set the KAGGLE_USERNAME and KAGGLE_KEY environment variables before running the next cell.
[ ]:
DATASET_ROOT = (Path(DOWNLOAD_PATH) / "sartorius-cell-instance-segmentation").resolve().absolute()
if not DATASET_ROOT.exists():
import zipfile
from kaggle import KaggleApi
api = KaggleApi()
api.authenticate()
print("Downloading dataset from Kaggle")
api.competition_download_files(
"sartorius-cell-instance-segmentation", path=Path(DOWNLOAD_PATH).absolute().as_posix()
)
with zipfile.ZipFile(f"{DOWNLOAD_PATH}/sartorius-cell-instance-segmentation.zip", "r") as zip_ref:
zip_ref.extractall(DATASET_ROOT)
else:
print(f"Dataset root {DATASET_ROOT} already exists")
Prepare the Table data¶
The annotations are stored in a csv file, with one row per instance.
We’ll read the csv file and group the annotations by image_id, then convert the instance annotations to COCO RLE format before writing to a Table.
[ ]:
train_csv_file = DATASET_ROOT / "train.csv"
assert train_csv_file.exists(), f"Train CSV file {train_csv_file} does not exist"
train_csv = pd.read_csv(train_csv_file)
train_csv.head()
[ ]:
# Map cell names to indices
cell_types_to_index = {"astro": 0, "cort": 1, "shsy5y": 2}
[ ]:
# Group annotations by image_id
image_annotations = {}
for _, row in tqdm(train_csv.iterrows(), total=len(train_csv), desc="Grouping annotations by image_id"):
image_id = row["id"]
if image_id not in image_annotations:
image_annotations[image_id] = {
"width": row["width"],
"height": row["height"],
"sample_id": row["sample_id"],
"annotations": [],
}
# Add this annotation
annotation = {
"cell_type_index": cell_types_to_index[row["cell_type"]],
"segmentation": list(map(int, row["annotation"].split())),
}
image_annotations[image_id]["annotations"].append(annotation)
[ ]:
def starts_lengths_to_coco_rle(starts_lengths, image_height, image_width):
"""Convert a list of starts and lengths to a COCO RLE by creating a binary mask and encoding it."""
# Convert to numpy array and get starts/lengths
s = np.array(starts_lengths, dtype=int)
starts = s[0::2] - 1 # Convert from 1-based to 0-based indexing
lengths = s[1::2]
# Create binary mask
mask = np.zeros(image_height * image_width, dtype=np.uint8)
for start, length in zip(starts, lengths):
mask[start : start + length] = 1
mask = mask.reshape(image_height, image_width)
# Convert to COCO RLE format
rle = mask_utils.encode(np.asfortranarray(mask))
return rle["counts"].decode("utf-8")
[ ]:
def annotations_to_3lc_format(image_annotations):
"""Convert a list of annotations to the format required by 3LC instance segmentation Tables.
Input format:
{
"cell_type_index": int,
"segmentation": list[int],
"width": int,
"height": int,
}
Output format:
{
"image_height": int,
"image_width": int,
"rles": list[bytes],
"instance_properties": {
"cell_type": list[int],
}
}
"""
image_height = image_annotations["height"]
image_width = image_annotations["width"]
rles = []
cell_types = []
for annotation in image_annotations["annotations"]:
rle = starts_lengths_to_coco_rle(annotation["segmentation"], image_height, image_width)
rles.append(rle)
cell_types.append(annotation["cell_type_index"])
return {
"image_height": image_height,
"image_width": image_width,
"rles": rles,
"instance_properties": {
"label": cell_types,
},
}
Now we can collect all the transformed column-data for the Table.
[ ]:
sample_ids = []
image_paths = []
segmentations = []
for image_id, image_data in tqdm(image_annotations.items(), total=len(image_annotations), desc="Processing images"):
sample_ids.append(image_data["sample_id"])
image_paths.append(
tlc.Url(DATASET_ROOT / "train" / f"{image_id}.png").to_relative().to_str()
) # Call to_relative() to ensure aliases are applied
segmentations.append(annotations_to_3lc_format(image_data))
Create Table¶
Create a Table using a TableWriter and a provided schema.
[ ]:
table_data = {
"sample_id": sample_ids,
"image": image_paths,
"segmentations": segmentations,
}
table_schemas = {
"image": tlc.ImageUrlSchema(sample_type="PILImage"),
"segmentations": tlc.SegmentationSchema(
label_value_map={v: tlc.MapElement(k) for k, v in cell_types_to_index.items()},
sample_type=tlc.InstanceSegmentationMasks.sample_type,
),
}
table = tlc.Table.from_dict(
table_data,
structure=table_schemas,
project_name=PROJECT_NAME,
dataset_name=DATASET_NAME,
table_name=TABLE_NAME,
if_exists="rename",
)
Plot a sample from the Table¶
Fetch the first sample from the Table, plot the image and the instance masks.
[ ]:
first_sample = table[0]
first_sample["image"]
[ ]:
# Plot the image path to ensure the alias is working
print(f"First sample image path: {table.table_rows[0]['image']}")
[ ]:
masks = first_sample["segmentations"]["masks"]
combined_mask = masks.sum(axis=2) > 0 # Combine all instance masks to a single mask for plotting
plt.imshow(combined_mask, cmap="gray")
plt.show()
Convert to polygons and make dataset splits¶
To end this example, we’ll convert the Table to a format compatible with YOLO and make train/val splits.
[ ]:
from tlc_tools.derived_tables import masks_to_polygons
# Creates an EditedTable where the sample type of the segmentation is changed from masks to polygons
polygon_table = masks_to_polygons(table)
[ ]:
first_sample = polygon_table[0]
polygons = first_sample["segmentations"]["polygons"]
fig, ax = plt.subplots()
for polygon in polygons:
vertices = np.array(polygon).reshape(-1, 2)
path = MatplotlibPath(vertices)
patch = patches.PathPatch(path, facecolor="#00FFFF", edgecolor="black")
ax.add_patch(patch)
# Set axis limits based on image dimensions
ax.set_xlim(0, first_sample["segmentations"]["image_width"])
ax.set_ylim(0, first_sample["segmentations"]["image_height"])
plt.show()
[ ]:
from tlc_tools.split import split_table
splits = split_table(polygon_table)