View source
Download
.ipynb
Fine-tune Hugging Face SegFormer on a custom dataset¶
This tutorial covers metrics collection on a custom semantic segmentation dataset using 3lc and training using 🤗 transformers.
It is based on the original notebook found here.
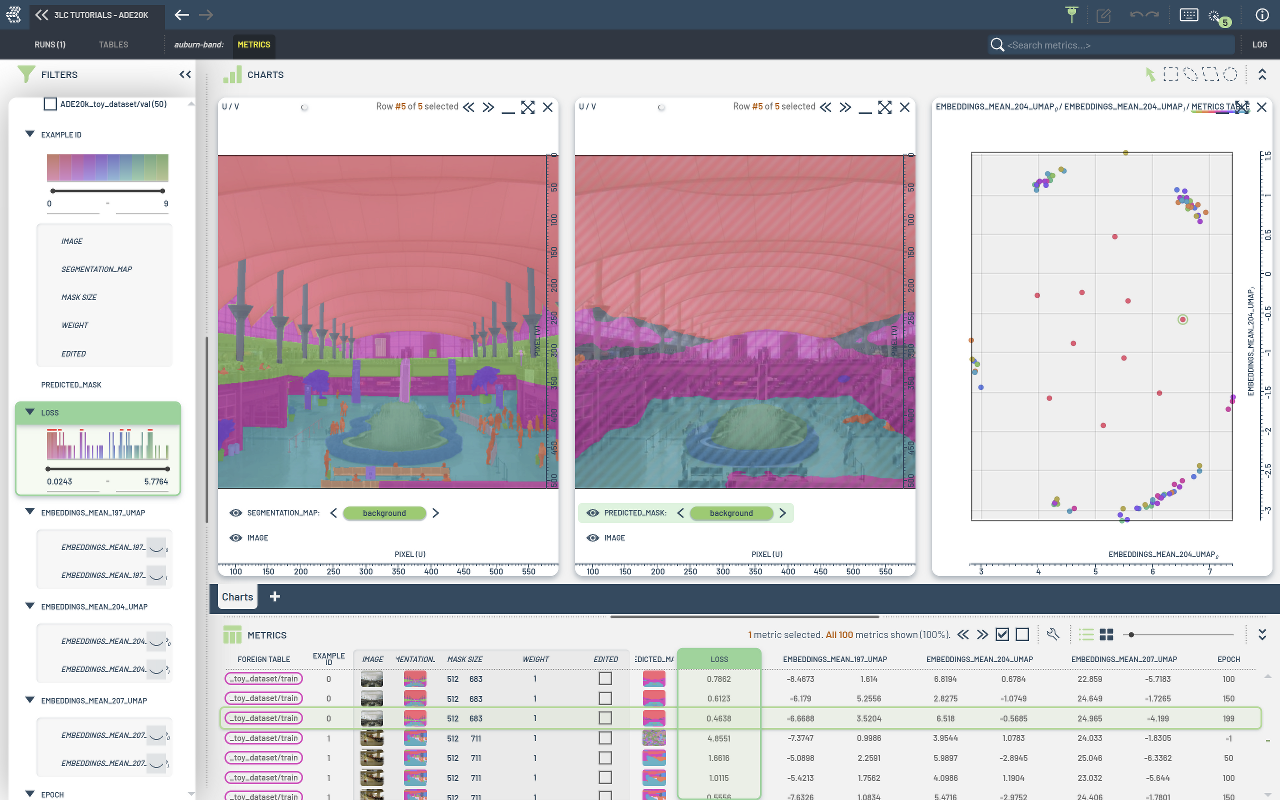
A small subset of the ADE20K dataset is used for this tutorial. The subset consists of 5 training images and 5 validation images, with semantic masks containing 150 labels.
During training, per-sample loss, embeddings, and predictions are collected.
Project setup¶
[ ]:
PROJECT_NAME = "3LC Tutorials - Semantic Segmentation ADE20k"
DATASET_NAME = "ADE20k_toy_dataset"
TMP_PATH = "../../transient_data"
EPOCHS = 200
NUM_WORKERS = 0
BATCH_SIZE = 2
Install dependencies¶
[ ]:
%pip install 3lc[huggingface] "transformers<=4.56.0"
%pip install git+https://github.com/3lc-ai/3lc-examples.git
Imports¶
[ ]:
import json
import os
from pathlib import Path
import tlc
import torch
from huggingface_hub import hf_hub_download
from PIL import Image
from torch.utils.data import DataLoader, Dataset
from tqdm.notebook import tqdm
from transformers import SegformerImageProcessor
from tlc_tools.common import download_and_extract_zipfile, infer_torch_device
Download the dataset¶
Fetch the label map from the Hugging Face Hub¶
[ ]:
# load id2label mapping from a JSON on the hub
repo_id = "huggingface/label-files"
filename = "ade20k-id2label.json"
with open(hf_hub_download(repo_id=repo_id, filename=filename, repo_type="dataset")) as f:
id2label = json.load(f)
id2label = {int(k): v for k, v in id2label.items()}
label2id = {v: k for k, v in id2label.items()}
unreduced_label_map = {0.0: "background", **{k + 1: v for k, v in id2label.items()}}
[ ]:
id2label
Initialize a Run¶
Setup Torch Datasets and 3LC Tables¶
[ ]:
class SemanticSegmentationDataset(Dataset):
"""Image (semantic) segmentation dataset."""
def __init__(self, root_dir: str, train: bool = True):
"""
:param root_dir: Root directory of the dataset containing the images + annotations.
:param train: Whether to load "training" or "validation" images + annotations.
"""
self.root_dir = root_dir
self.train = train
sub_path = "training" if self.train else "validation"
self.img_dir = os.path.join(self.root_dir, "images", sub_path)
self.ann_dir = os.path.join(self.root_dir, "annotations", sub_path)
# read images
image_file_names = []
for _, _, files in os.walk(self.img_dir):
image_file_names.extend(files)
self.images = sorted(image_file_names)
# read annotations
annotation_file_names = []
for _, _, files in os.walk(self.ann_dir):
annotation_file_names.extend(files)
self.annotations = sorted(annotation_file_names)
assert len(self.images) == len(self.annotations), "There must be as many images as there are segmentation maps"
def __len__(self):
return len(self.images)
def __getitem__(self, idx):
image = Image.open(os.path.join(self.img_dir, self.images[idx]))
segmentation_map = Image.open(os.path.join(self.ann_dir, self.annotations[idx]))
# We need to include the original segmentation map size, in order to post-process the model output
return image, segmentation_map, (segmentation_map.size[1], segmentation_map.size[0])
[ ]:
train_dataset = SemanticSegmentationDataset(root_dir=DATASET_ROOT, train=True)
val_dataset = SemanticSegmentationDataset(root_dir=DATASET_ROOT, train=False)
[ ]:
train_dataset[0][1]
Create the Tables¶
[ ]:
structure = (
tlc.PILImage("image"),
tlc.SegmentationPILImage("segmentation_map", classes=unreduced_label_map),
tlc.HorizontalTuple("mask size", [tlc.Int("width"), tlc.Int("height")]),
)
train_table = tlc.Table.from_torch_dataset(
train_dataset,
structure,
project_name=PROJECT_NAME,
dataset_name=DATASET_NAME,
table_name="train",
if_exists="overwrite",
)
val_table = tlc.Table.from_torch_dataset(
val_dataset,
structure,
project_name=PROJECT_NAME,
dataset_name=DATASET_NAME,
table_name="val",
if_exists="overwrite",
)
[ ]:
class MapFn:
def __init__(self, image_processor: SegformerImageProcessor):
self.image_processor = image_processor
def __call__(self, sample):
image, segmentation_map, mask_size = sample
encoded_inputs = self.image_processor(image, segmentation_map, return_tensors="pt")
for k, _ in encoded_inputs.items():
encoded_inputs[k].squeeze_() # remove batch dimension
encoded_inputs.update({"mask_size": torch.tensor(mask_size)})
return encoded_inputs
image_processor = SegformerImageProcessor(reduce_labels=True)
# Apply the image processor to the datasets
train_table.map(MapFn(image_processor))
val_table.map(MapFn(image_processor))
[ ]:
train_table[0].keys()
[ ]:
train_table.url
Define the model¶
[ ]:
from transformers import SegformerForSemanticSegmentation
model = SegformerForSemanticSegmentation.from_pretrained(
"nvidia/mit-b0",
num_labels=150,
id2label=id2label,
label2id=label2id,
).to(DEVICE)
[ ]:
# Predict on single sample
model(train_table[0]["pixel_values"].unsqueeze(0).to(DEVICE))
Setup metrics collection¶
[ ]:
# 1. EmbeddingsMetricsCollector to collect hidden layer activations
for ind, layer in enumerate(model.named_modules()):
print(ind, "=>", layer[0])
# Interesting layers for embedding collection:
# - segformer.encoder.layer_norm.3 (Index: 197)
# - decode_head.linear_c.2.proj (Index: 204)
# - decode_head.linear_c.3.proj (Index: 207)
layers = [197, 204, 207]
embedding_collector = tlc.EmbeddingsMetricsCollector(layers=layers)
[ ]:
# 2. A metrics collection callable to collect per-sample loss
def metrics_fn(batch, predictor_output):
labels = batch["labels"].to(DEVICE)
logits = predictor_output.forward.logits
upsampled_logits = torch.nn.functional.interpolate(
logits, size=labels.shape[-2:], mode="bilinear", align_corners=False
)
loss = torch.nn.functional.cross_entropy(upsampled_logits, labels, reduction="none", ignore_index=255)
loss = loss.mean(dim=(1, 2))
return {"loss": loss.detach().cpu().numpy()}
[ ]:
# 3. A SegmentationMetricsCollector to write out the predictions
def preprocess_fn(batch, predictor_output: tlc.PredictorOutput):
"""Convert logits to masks with the same size as the input, un-reduce the labels"""
processed_masks = image_processor.post_process_semantic_segmentation(
predictor_output.forward,
batch["mask_size"].tolist(),
)
for i in range(len(processed_masks)):
mask = processed_masks[i]
mask[mask == 255] = 0
mask = mask + 1
processed_masks[i] = mask
return batch, processed_masks
segmentation_collector = tlc.SegmentationMetricsCollector(label_map=unreduced_label_map, preprocess_fn=preprocess_fn)
[ ]:
# Define a single function to collect all metrics
# A Predictor object wraps the model and enables embedding-collection
predictor = tlc.Predictor(model, device=DEVICE, layers=layers)
# Control the arguments used for the dataloader used during metrics collection
mc_dataloader_args = {"batch_size": BATCH_SIZE}
def collect_metrics(epoch):
tlc.collect_metrics(
train_table,
[segmentation_collector, metrics_fn, embedding_collector],
predictor,
constants={"epoch": epoch},
dataloader_args=mc_dataloader_args,
split="train",
)
tlc.collect_metrics(
val_table,
[segmentation_collector, metrics_fn, embedding_collector],
predictor,
constants={"epoch": epoch},
dataloader_args=mc_dataloader_args,
split="val",
)
# Collect metrics before training (-1 means before training)
collect_metrics(-1)
Train!¶
[ ]:
# Uses the "weights" column of the Table to sample the data
sampler = train_table.create_sampler()
train_dataloader = DataLoader(train_table, batch_size=BATCH_SIZE, sampler=sampler, num_workers=NUM_WORKERS)
valid_dataloader = DataLoader(val_table, batch_size=BATCH_SIZE, num_workers=NUM_WORKERS)
[ ]:
def loss_fn(logits, labels):
upsampled_logits = torch.nn.functional.interpolate(
logits, size=labels.shape[-2:], mode="bilinear", align_corners=False
)
if model.config.num_labels > 1:
loss_fct = torch.nn.CrossEntropyLoss(ignore_index=model.config.semantic_loss_ignore_index)
loss = loss_fct(upsampled_logits, labels)
elif model.config.num_labels == 1:
valid_mask = ((labels >= 0) & (labels != model.config.semantic_loss_ignore_index)).float()
loss_fct = torch.nn.BCEWithLogitsLoss(reduction="none")
loss = loss_fct(upsampled_logits.squeeze(1), labels.float())
loss = (loss * valid_mask).mean()
return loss
[ ]:
# define optimizer
optimizer = torch.optim.AdamW(model.parameters(), lr=0.00006)
# move model to GPU
model.to(DEVICE)
model.train()
for epoch in range(EPOCHS): # loop over the dataset multiple times
print("Epoch:", epoch)
agg_loss = 0.0
seen_samples = 0
for _idx, batch in enumerate(tqdm(train_dataloader)):
# get the inputs;
pixel_values = batch["pixel_values"].to(DEVICE)
labels = batch["labels"].to(DEVICE)
# zero the parameter gradients
optimizer.zero_grad()
# forward + backward + optimize
outputs = model(pixel_values=pixel_values, labels=labels)
_, logits = outputs.loss, outputs.logits
loss = loss_fn(outputs.logits, labels)
agg_loss += loss.item() * pixel_values.shape[0]
seen_samples += pixel_values.shape[0]
loss.backward()
optimizer.step()
# evaluate
with torch.no_grad():
upsampled_logits = torch.nn.functional.interpolate(
logits, size=labels.shape[-2:], mode="bilinear", align_corners=False
)
predicted = upsampled_logits.argmax(dim=1)
# Log aggregated metrics directly to the active Run
tlc.log(
{
"epoch": epoch,
"running_train_loss": loss.item() / seen_samples,
}
)
if epoch % 50 == 0 and epoch != 0:
collect_metrics(epoch)
Collect metrics after training¶
[ ]:
collect_metrics(epoch)
Dimensionality reduce collected metrics¶
[ ]:
run.reduce_embeddings_by_foreign_table_url(train_table.url)