View source
Download
.ipynb
Train autoencoder for embedding extraction¶
This notebook showcases more ways of working with metrics and embeddings in 3LC. It is mostly meant as a demonstration of how to collect embeddings and image metrics from a manually trained model.
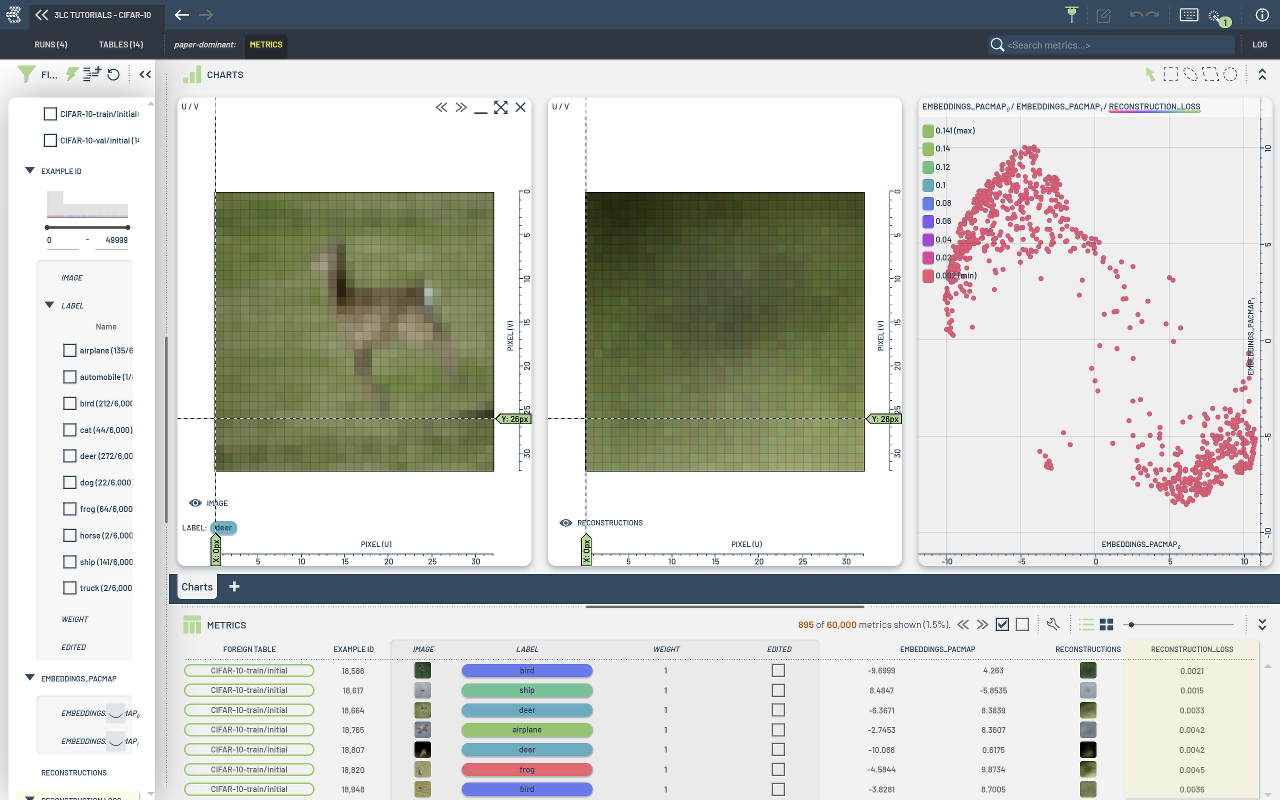
The auto-encoder architecture is mainly used as a simple example to demonstrate the process, and the model should only be considered as an example of an embedding extractor, which also produces images as a side effect.
Install dependencies¶
[ ]:
%pip install 3lc[pacmap]
%pip install git+https://github.com/3lc-ai/3lc-examples.git
%pip install timm
Imports¶
[ ]:
import tlc
import torch
import torch.nn as nn
from timm import create_model
from torch.utils.data import DataLoader
from torchvision import transforms
from tqdm.auto import tqdm
from tlc_tools.common import infer_torch_device
Project setup¶
[ ]:
TMP_PATH = "../../transient_data"
PROJECT_NAME = "3LC Tutorials - CIFAR-10"
RUN_NAME = "Train autoencoder"
RUN_DESCRIPTION = "Train an autoencoder and collect embeddings and reconstructions"
BACKBONE = "resnet50"
EMBEDDING_DIM = 512 # Desired embedding dimension
EPOCHS = 10
FREEZE_ENCODER = False
IMAGE_WIDTH = 32
IMAGE_HEIGHT = 32
NUM_CHANNELS = 3
BATCH_SIZE = 64
METHOD = "pacmap"
NUM_COMPONENTS = 2
NUM_WORKERS = 0
[ ]:
CHECKPOINT_PATH = TMP_PATH + "/autoencoder_model.pth"
Load input Table¶
[ ]:
train_table = tlc.Table.from_names("initial", "CIFAR-10-train", "3LC Tutorials - CIFAR-10")
val_table = tlc.Table.from_names("initial", "CIFAR-10-val", "3LC Tutorials - CIFAR-10")
[ ]:
# Prepare Data
transform = transforms.Compose(
[
transforms.ToTensor(),
]
)
def map_fn(sample):
"""Map samples from the table to be compatible with the model."""
image = sample[0]
image = transform(image)
return image
train_table.clear_maps()
train_table.map(map_fn)
val_table.clear_maps()
val_table.map(map_fn)
Train autoencoder¶
[ ]:
class Autoencoder(nn.Module):
def __init__(self, backbone_name="resnet50", embedding_dim=512, freeze_encoder=FREEZE_ENCODER):
super().__init__()
# Load the backbone as an encoder
self.encoder = create_model(backbone_name, pretrained=True, num_classes=0)
encoder_output_dim = self.encoder.feature_info[-1]["num_chs"]
# Freeze encoder parameters if specified
if freeze_encoder:
for param in self.encoder.parameters():
param.requires_grad = False
# Add a projection layer to reduce to embedding_dim
self.projector = nn.Linear(encoder_output_dim, embedding_dim)
# Define the decoder
self.decoder = nn.Sequential(
nn.Linear(embedding_dim, encoder_output_dim),
nn.ReLU(),
nn.Linear(encoder_output_dim, IMAGE_HEIGHT * IMAGE_WIDTH * NUM_CHANNELS),
nn.Sigmoid(),
)
def forward(self, x):
# Encoder
features = self.encoder(x)
embeddings = self.projector(features)
# Decoder
reconstructions = self.decoder(embeddings)
reconstructions = reconstructions.view(x.size(0), NUM_CHANNELS, IMAGE_WIDTH, IMAGE_HEIGHT)
return embeddings, reconstructions
[ ]:
# Initialize the model
model = Autoencoder(backbone_name=BACKBONE, embedding_dim=EMBEDDING_DIM)
# Training Components
criterion = nn.MSELoss() # Reconstruction loss
optimizer = torch.optim.Adam(model.parameters(), lr=1e-4)
# Create data loaders
train_loader = DataLoader(train_table, batch_size=BATCH_SIZE, shuffle=True)
val_loader = DataLoader(val_table, batch_size=BATCH_SIZE, shuffle=False)
device = infer_torch_device()
model.to(device)
[ ]:
# Training loop
for epoch in range(EPOCHS):
model.train()
epoch_train_loss = 0.0
epoch_val_loss = 0.0
for images in tqdm(train_loader, desc="Training", total=len(train_loader)):
images = images.to(device)
# Forward pass
embeddings, reconstructions = model(images)
# Compute loss
loss = criterion(reconstructions, images)
# Backward pass and optimization
optimizer.zero_grad()
loss.backward()
optimizer.step()
epoch_train_loss += loss.item()
# Validation pass
model.eval()
with torch.no_grad():
for images in tqdm(val_loader, desc="Validation", total=len(val_loader)):
images = images.to(device)
# Forward pass
embeddings, reconstructions = model(images)
# Compute loss
loss = criterion(reconstructions, images)
epoch_val_loss += loss.item()
epoch_train_loss /= len(train_loader)
epoch_val_loss /= len(val_loader)
print(f"Epoch [{epoch + 1}/{EPOCHS}], Train Loss: {epoch_train_loss:.4f}, Val Loss: {epoch_val_loss:.4f}")
[ ]:
# Save the model
torch.save(model.state_dict(), CHECKPOINT_PATH)
print(f"Model saved to {CHECKPOINT_PATH}")
Collect metrics from the trained model¶
[ ]:
unreduced_loss = nn.MSELoss(reduction="none") # Reconstruction loss
def metrics_fn(batch, predictor_output):
embeddings, reconstructions = predictor_output.forward
reconstructed_images = [transforms.ToPILImage()(image.cpu()) for image in reconstructions]
reconstruction_loss = unreduced_loss(reconstructions.to(device), batch.to(device)).mean(dim=(1, 2, 3))
return {
"embeddings": embeddings.cpu().detach().numpy(),
"reconstructions": reconstructed_images,
"reconstruction_loss": reconstruction_loss.cpu().detach().numpy(),
}
[ ]:
run = tlc.init(project_name=PROJECT_NAME, run_name=RUN_NAME, description=RUN_DESCRIPTION)
tlc.collect_metrics(
train_table,
metrics_fn,
model,
collect_aggregates=False,
dataloader_args={"batch_size": BATCH_SIZE, "num_workers": NUM_WORKERS},
)
tlc.collect_metrics(
val_table,
metrics_fn,
model,
collect_aggregates=False,
dataloader_args={"batch_size": BATCH_SIZE, "num_workers": NUM_WORKERS},
)
Reduce embeddings to 2D¶
[ ]:
run.reduce_embeddings_by_foreign_table_url(
train_table.url,
source_embedding_column="embeddings",
method=METHOD,
n_components=NUM_COMPONENTS,
)