View source
Download
## Visualize Sample Results
.ipynb
Auto-segment images using SAM¶
This notebook creates a 3LC Table with auto-generated segmentation masks from SAM using grid-based point prompting.
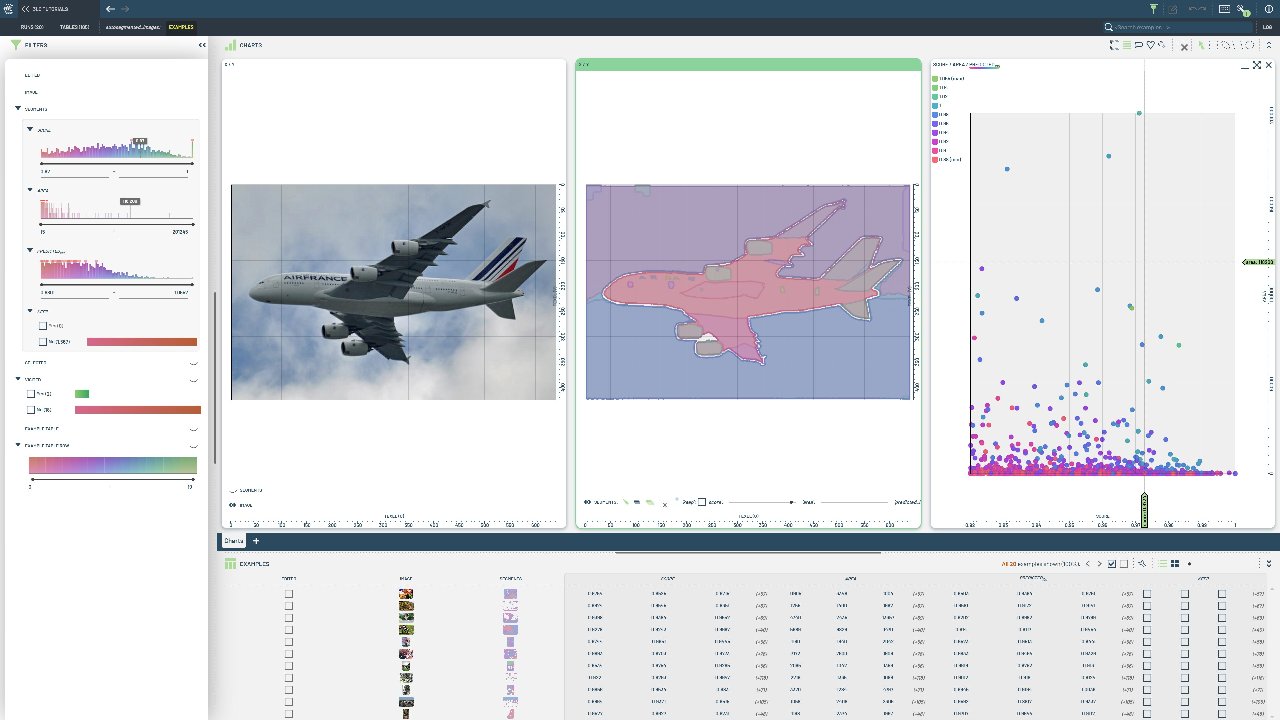
Unlike the bounding box-based approach, this method automatically discovers objects in images without requiring ground truth annotations.
Install dependencies¶
[ ]:
%pip install 3lc
%pip install git+https://github.com/3lc-ai/3lc-examples.git
%pip install git+https://github.com/facebookresearch/segment-anything
%pip install matplotlib
Imports¶
Project Setup¶
[ ]:
PROJECT_NAME = "3LC Tutorials - COCO128"
DATASET_NAME = "AutoSegmented Images"
TABLE_NAME = "autosegmented_images"
MODEL_TYPE = "vit_b"
TMP_PATH = "../../transient_data"
# Image dataset configuration
DATA_PATH = "../../data"
MAX_IMAGES = 20 # Limit for initial testing - set to None for all images
# Segmentation filtering parameters
MIN_AREA_THRESHOLD = 1000 # Minimum area in pixels to keep a segment
MIN_CONFIDENCE_THRESHOLD = 0.7 # Minimum confidence score to keep a segment
POINTS_PER_SIDE = 32
PRED_IOU_THRESHOLD = 0.88
Load Images¶
[ ]:
# Get list of image files
image_dir = (Path(DATA_PATH) / "coco128" / "images").resolve()
image_extensions = {".jpg", ".jpeg", ".png", ".bmp", ".tiff"}
image_files = [f for f in image_dir.glob("*") if f.suffix.lower() in image_extensions]
if MAX_IMAGES is not None:
image_files = image_files[:MAX_IMAGES]
print(f"Found {len(image_files)} images to process")
print(f"Sample images: {[f.name for f in image_files[:5]]}")
Initialize SAM Automatic Mask Generator¶
We’ll use SAM’s SamAutomaticMaskGenerator which uses automatic point grid prompting to segment all objects in each image without requiring any input prompts.
[ ]:
# Download checkpoint
model_url = "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_b_01ec64.pth"
# Download the SAM model checkpoint if it doesn't already exist
checkpoint_path = Path(TMP_PATH) / "sam_vit_b_01ec64.pth"
if not checkpoint_path.exists():
print(f"Downloading SAM checkpoint to {checkpoint_path}...")
checkpoint_path.parent.mkdir(parents=True, exist_ok=True)
response = requests.get(model_url, stream=True)
total_size = int(response.headers.get("content-length", 0))
with (
open(checkpoint_path, "wb") as f,
tqdm(
desc="Downloading",
total=total_size,
unit="B",
unit_scale=True,
unit_divisor=1024,
) as bar,
):
for chunk in response.iter_content(chunk_size=8192):
if chunk:
f.write(chunk)
bar.update(len(chunk))
print("Download completed.")
else:
print(f"Checkpoint already exists at {checkpoint_path}.")
[ ]:
# Load SAM model
device = infer_torch_device()
sam = sam_model_registry[MODEL_TYPE](checkpoint=checkpoint_path)
sam.to(device=device)
# Initialize the automatic mask generator
mask_generator = SamAutomaticMaskGenerator(
model=sam,
points_per_side=POINTS_PER_SIDE, # Grid size for point prompts
pred_iou_thresh=PRED_IOU_THRESHOLD, # IoU threshold for mask quality filtering
stability_score_thresh=0.92, # Stability score threshold
crop_n_layers=1, # Number of crop layers
crop_n_points_downscale_factor=2, # Downscale factor for crop points
min_mask_region_area=MIN_AREA_THRESHOLD, # Minimum area in pixels
)
print(f"Initialized SAM Automatic Mask Generator on device: {device}")
print(f"Using model type: {MODEL_TYPE}")
print(f"Checkpoint: {checkpoint_path}")
Process Images and Generate Masks¶
[ ]:
# Process each image and collect segmentations
segmentations_data = []
for image_path in tqdm(image_files, desc="Processing images", total=len(image_files)):
# Load image
image = cv2.imread(str(image_path))
image_rgb = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# Generate masks for this image
masks = mask_generator.generate(image_rgb)
if masks:
# Convert masks to the format expected by 3LC
h, w = image_rgb.shape[:2]
# Stack all masks for this image into a single array
mask_array = np.stack([mask["segmentation"] for mask in masks], axis=2).astype(np.uint8)
# Create instance properties (scores, areas, etc.)
instance_properties = {
"score": [mask["stability_score"] for mask in masks],
"area": [mask["area"] for mask in masks],
"predicted_iou": [mask["predicted_iou"] for mask in masks],
"keep": [False] * len(masks),
}
segmentation_data = {
"image_height": h,
"image_width": w,
"masks": mask_array,
"instance_properties": instance_properties,
}
row_data = {
"image": tlc.Url(image_path).to_relative().to_str(),
"segments": segmentation_data,
}
segmentations_data.append(row_data)
print(f"\\nProcessed {len(segmentations_data)} images with valid segmentations")
Create 3LC Table¶
[ ]:
# Create the 3LC table with auto-generated segmentations
table_writer = tlc.TableWriter(
project_name=PROJECT_NAME,
dataset_name=DATASET_NAME,
table_name=TABLE_NAME,
column_schemas={
"image": tlc.ImageUrlSchema(),
"segments": tlc.InstanceSegmentationMasks(
"segments",
instance_properties_structure={
"score": tlc.Schema(value=tlc.Float32Value(0, 1), writable=False),
"area": tlc.Schema(value=tlc.Int32Value(), writable=False),
"predicted_iou": tlc.Schema(value=tlc.Float32Value(0, 1), writable=False),
"keep": tlc.Schema(value=tlc.BoolValue(), writable=True),
},
),
},
)
# Add all the segmentation data to the table
for row_data in segmentations_data:
table_writer.add_row(row_data)
# Finalize the table
table = table_writer.finalize()
print(f"\nCreated 3LC table: {table.name}")
print(f"Table URL: {table.url}")
print(f"Total rows: {len(table)}")
# Display some statistics
total_segments = sum(len(row["segments"]["instance_properties"]["score"]) for row in segmentations_data)
avg_segments_per_image = total_segments / len(segmentations_data) if segmentations_data else 0
print("\nSegmentation Statistics:")
print(f"Total segments collected: {total_segments}")
print(f"Average segments per image: {avg_segments_per_image:.1f}")
[ ]:
import matplotlib.pyplot as plt
if len(table) > 0:
# Get a sample from the table
sample_idx = 0
sample = table[sample_idx]
# Load the original image
image_path = sample["image"]
image = cv2.imread(image_path)
image_rgb = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# Get the segmentation masks
masks = sample["segments"]["masks"]
scores = sample["segments"]["instance_properties"]["score"]
areas = sample["segments"]["instance_properties"]["area"]
print(f"Sample image: {Path(image_path).name}")
print(f"Number of segments: {masks.shape[2]}")
print(f"Score range: {min(scores):.3f} - {max(scores):.3f}")
print(f"Area range: {min(areas)} - {max(areas)} pixels")
# Create visualization
fig, axes = plt.subplots(1, 3, figsize=(18, 6))
# Original image
axes[0].imshow(image_rgb)
axes[0].set_title("Original Image")
axes[0].axis("off")
# All masks overlay
axes[1].imshow(image_rgb)
combined_mask = np.zeros((masks.shape[0], masks.shape[1]))
colors = plt.cm.tab20(np.linspace(0, 1, min(20, masks.shape[2])))
for i in range(min(masks.shape[2], 20)): # Show up to 20 masks
mask = masks[:, :, i]
if mask.sum() > 0:
# Create colored overlay
colored_mask = np.zeros((*mask.shape, 4))
colored_mask[mask == 1] = colors[i % len(colors)]
axes[1].imshow(colored_mask, alpha=0.7)
axes[1].set_title(f"All Segments Overlay ({min(masks.shape[2], 20)} shown)")
axes[1].axis("off")
# Individual high-quality masks
axes[2].imshow(image_rgb)
# Show only the top 5 masks by score
top_indices = sorted(range(len(scores)), key=lambda i: scores[i], reverse=True)[:5]
for i, mask_idx in enumerate(top_indices):
mask = masks[:, :, mask_idx]
if mask.sum() > 0:
colored_mask = np.zeros((*mask.shape, 4))
colored_mask[mask == 1] = colors[i % len(colors)]
axes[2].imshow(colored_mask, alpha=0.8)
axes[2].set_title("Top 5 Segments by Score")
axes[2].axis("off")
plt.tight_layout()
plt.show()
else:
print("No images processed successfully")