.ipynb
Apply dimensionality reduction to multiple Tables¶
This example shows how to use the “producer-consumer” pattern for re-using dimensionality reduction models across different tables.
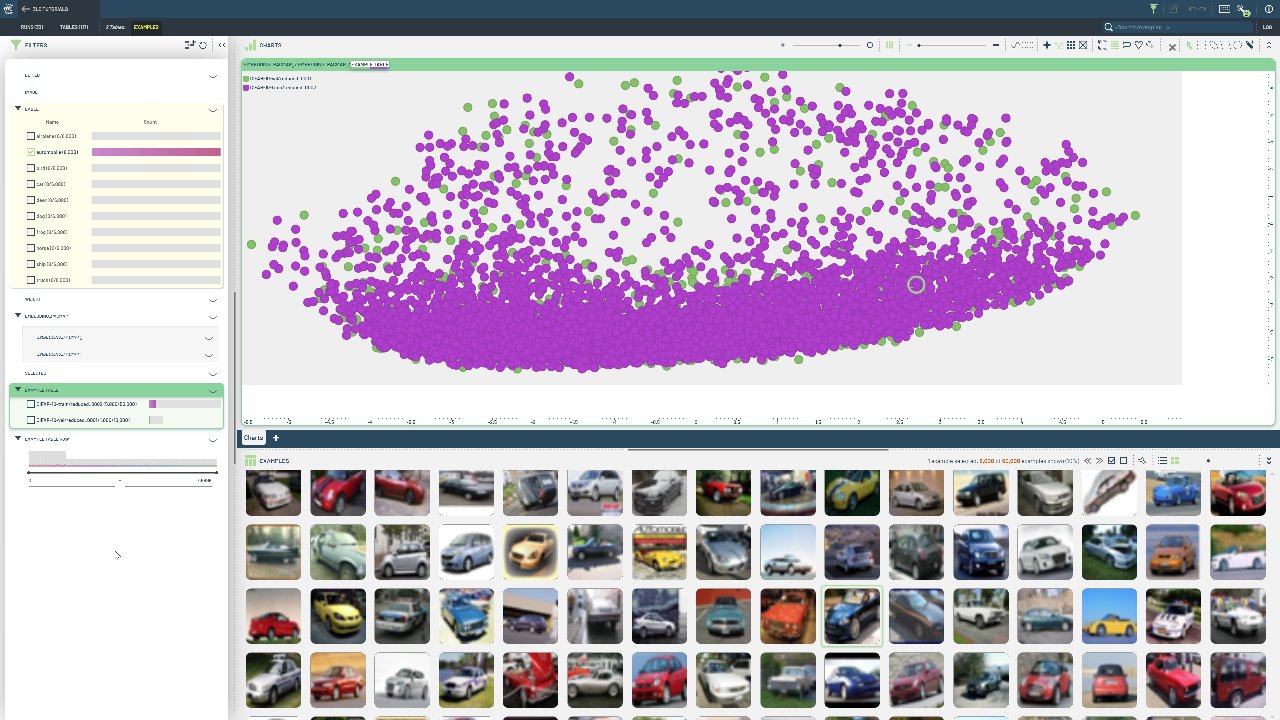
Specifically, high-dimensional embeddings from the same model are added as new columns to the train and val split of the CIFAR-10 dataset. With a single call, a UMAP model is trained on the train split embeddings, and then used to transform both the train and val split embeddings. This ensures that the reduced, 3-dimensional embeddings are mapped to the same space, which is crucial for comparing embeddings across tables.
The tlc package contains several helper functions for working with dimensionality reduction, and currently support both the UMAP and PaCMAP algorithms. A “producer” table is a reduction table that fits a dimensionality reduction model to the data, and saves the model for later use. A “consumer” table is a reduction table that uses the model from a producer table to only transform the data.
Project setup¶
[ ]:
PROJECT_NAME = "3LC Tutorials - CIFAR-10"
MODEL_NAME = "resnet18"
METHOD = "pacmap"
BATCH_SIZE = 32
DOWNLOAD_PATH = "../../transient_data"
NUM_COMPONENTS = 2
Install dependencies¶
[ ]:
%pip install 3lc[huggingface,pacmap]
%pip install timm
%pip install git+https://github.com/3lc-ai/3lc-examples
Imports¶
Load input Tables¶
We will re-use the CIFAR-10 tables created in an earlier notebook.
[ ]:
train_table = tlc.Table.from_names("initial", "CIFAR-10-train", PROJECT_NAME)
val_table = tlc.Table.from_names("initial", "CIFAR-10-val", PROJECT_NAME)
Load model¶
[ ]:
model = timm.create_model(MODEL_NAME, pretrained=True, num_classes=0, cache_dir=Path(DOWNLOAD_PATH) / "models")
model = model.to(infer_torch_device())
[ ]:
# Map the table to ensure only suitably preprocessed images are passed to the model
transform = Compose([Resize(256), ToTensor(), Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])])
def transformed_image(sample):
return transform(sample[0])
train_table.map(transformed_image)
val_table.map(transformed_image)
train_table[0]
[ ]:
train_table_with_embeddings = add_embeddings_to_table(table=train_table, model=model, batch_size=BATCH_SIZE)
[ ]:
val_table_with_embeddings = add_embeddings_to_table(table=val_table, model=model, batch_size=BATCH_SIZE)
Perform dimensionality reduction¶
[ ]:
url_mapping = tlc.reduce_embeddings_with_producer_consumer(
producer=val_table_with_embeddings,
consumers=[train_table_with_embeddings],
method=METHOD,
n_components=NUM_COMPONENTS,
)
[ ]:
reduced_train_table_url = url_mapping[train_table_with_embeddings.url]
reduced_val_table_url = url_mapping[val_table_with_embeddings.url]